Transcriptomics
Transcriptomics is the study of the transcriptome, which represents the complete set of RNA transcripts (mRNA, microRNA and other small RNAs) produced by the genome under specific conditions, at a particular time, in a particular cell or tissue. It provides a snapshot of gene expression levels and patterns, allowing researchers to understand how genes are activated or suppressed in response to various internal and external stimuli. Modern high-throughput techniques allow for the comprehensive analysis of multiple transcripts and their expression levels, allowing us to investigate the molecular mechanisms disease states. Various platforms are utilized for transcriptome analysis including Illumina short read bulkRNA sequencing, Oxford Nanopore Technologies long read sequencing or 10x Genomics for more specific analysis.
RNA sequencing (RNA-seq)
RNA-seq, a powerful tool utilizing Next-Generation Sequencing (NGS) technologies to generate comprehensive mapping and quantification of the transcriptome. It uncovers critical information, including gene expression levels within a cell, reflecting gene activity under a specific state such as disease development stages or other physiological states. Understanding the transcriptome is essential for interpreting genome functions and deciphering biological processes. Key goals of transcriptomics include cataloging all transcripts, defining gene function, and quantifying transcript expression under different conditions. RNA-Seq can also be utilized to detect novel transcripts without a reference genome, enabling de novo assembly of unexplored transcriptomes.
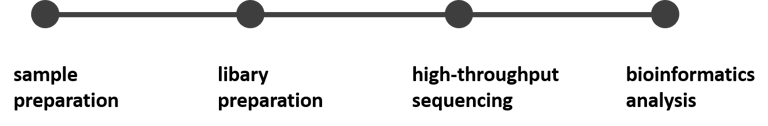

RNA-seq workflow
Gene expression profiling
Gene expression profiling is a cost-effective method for analyzing mRNA levels across different experimental conditions, helping to determine which genes are active ("on") or inactive ("off"). This technique reveals a cell's functional state and how it responds to treatments, environmental changes, or genetic variations. By comparing expression profiles, researchers can identify key genes linked to specific behaviors or conditions, helping understand disease mechanisms and therapeutic targets.
Single-cell sequencing
Single-cell RNA sequencing (scRNA-seq) is a state-of-the-art technique for analyzing gene expression at the individual cell level, allowing researchers to explore the heterogeneity and complexity of transcripts within cells. This method enables the identification of rare but functionally significant cell populations in both health and disease, addressing experimental questions that bulk analysis cannot resolve. By revealing cellular diversity, scRNA-seq provides critical insights into cellular functions, developmental processes, and interactions within microenvironments. Additionally, it supports personalized medicine by linking transcriptomic data to functional outcomes. When combined with other omics technologies, scRNA-seq offers a comprehensive understanding of cellular dynamics and regulatory mechanisms.


Sample types for RNA-seq




cell lines
tissue
lysates, biological fluids
extracellular vesicles
microbial cultures


plant material


