Proteomics
Proteomics is the large-scale study of proteomes, the complete set of proteins produced by an organism, tissue, or cell under specific conditions. The proteome is dynamic varying between cells and changes over time, influenced not only by gene expression but also by factors like post-translational modifications. Proteomics examines protein structure, function, interactions, and expression levels, offering insights into the pathogenesis of infection disease, host immune response and plant physiology. Key challenges in proteomics include identifying, quantifying, and characterizing proteins in complex systems, with emerging fields like spatial and single-cell proteomics providing deeper functional insights.
Discovery proteomics
The large-scale analysis of specific proteomes, is critical to the success of drug discovery, biomarker identification, and disease research. By analyzing protein diversity, abundances, modifications, and interactions, proteomics reveals the biological mechanisms underlying various organisms and diseases. Discovery proteomics, or untargeted proteomics, allows scientists to explore a proteome to identify proteins and proteoforms that control key biological processes. This approach is invaluable for gaining insights into everything from cell biology to infectious microbes to plant physiology.
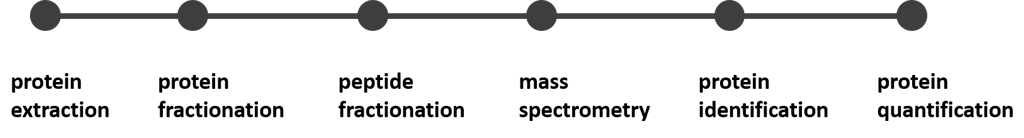
Proteomics workflow
Shotgun proteomics
This method uses the 'bottom-up' proteomics approach, proteins are identified by digesting them with trypsin and analyzing the resulting peptides using liquid chromatography-mass spectrometry (LC-MS). This method can be applied to individual proteins, such as those from SDS-PAGE gel fragments, or to a large number of proteins (hundreds to thousands) from whole cell lysates.
Quantitative proteomics
This is a powerful method employed in both discovery and targeted proteomic studies to explore and understand the overall dynamics of proteins within a cell, tissue, or organism. Several approaches, including protein mass spectrometry, are available to quantify relative changes in protein levels between samples, either by using isotopic labeling (eg. SILAC, TMT) or through label-free methods (eg. LFQ, DIA). Isotopic labeling can be carried out chemically at the peptide level or through the metabolic labeling of cells. Additionally, isotopically labeled standards can be used to measure the absolute concentration of proteins in a sample.

Targated proteomics
Targeted proteomics experiments are typically focused on quantifying fewer than 100 proteins with exceptional precision, sensitivity, specificity, and throughput. This approach often streamlines sample preparation to enhance both precision and efficiency. Targeted mass spectrometry quantitation employs specialized workflows and instruments designed to improve specificity and accurate measurement of a limited set of features across large sample sets. This can include techniques such as directed sequencing using inclusion lists and selected or multiple reaction monitoring. By restricting the number of monitored features, targeted proteomics optimizes chromatography, instrument tuning, and acquisition methods to achieve maximum sensitivity.
Sample types for proteomics




cell lines
tissue
lysates, biological fluids
extracellular vesicles
microbial cultures


plant materials


